Table of Contents
Team composition
| Name | |
| Robert Calef | rcalef@ucsc.edu |
| Chris Eisenhart | ceisenha@ucsc.edu |
| Natasha Dudek | natasha@dudek.org |
| Gepoliano Chaves | gchaves@ucsc.edu |
Discovar de novo overview
Discovar de novo is a next generation sequence assembly program. The program was developed by the Broad Institute and was released late in 2014. Discovar de novo is designed for 250 bp long illumina reads with the PCR duplicates and adaptor sequences removed. The following webpage contains the manual as provided by the Broad Institute (http://www.broadinstitute.org/software/discovar/blog/):
Team workflow
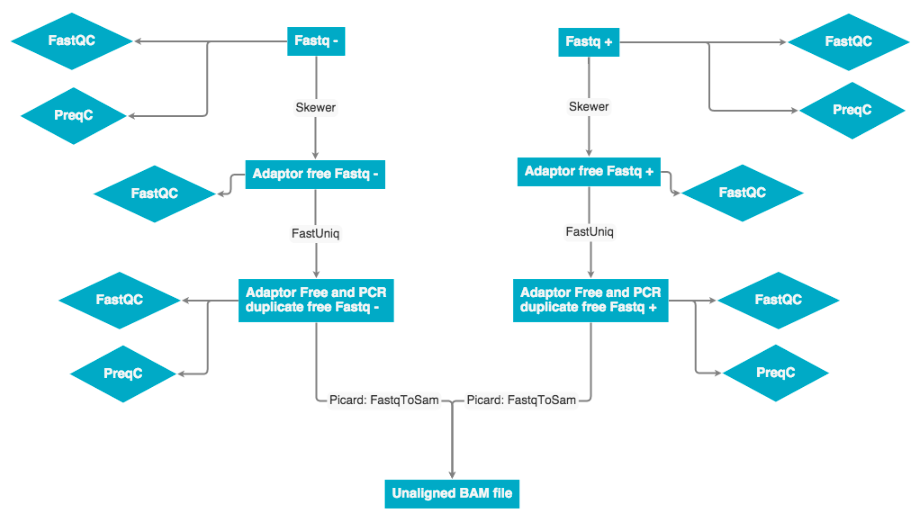
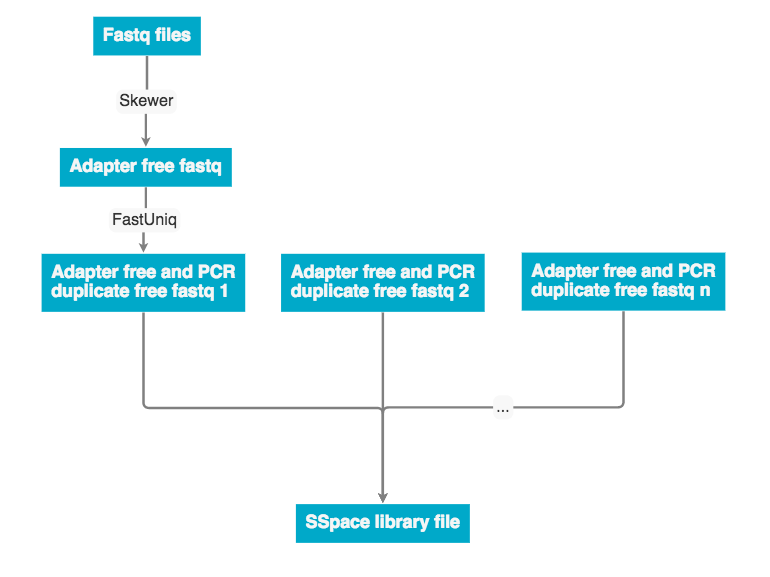
The raw data was received as fastq pairs. Each pair contains a forward and reverse strand. These pairs are ran through Skewer to remove adaptor sequences, then ran through fastUniq to remove PCR duplicates. Next the forward and reverse strand are merged into a single unaligned BAM file.
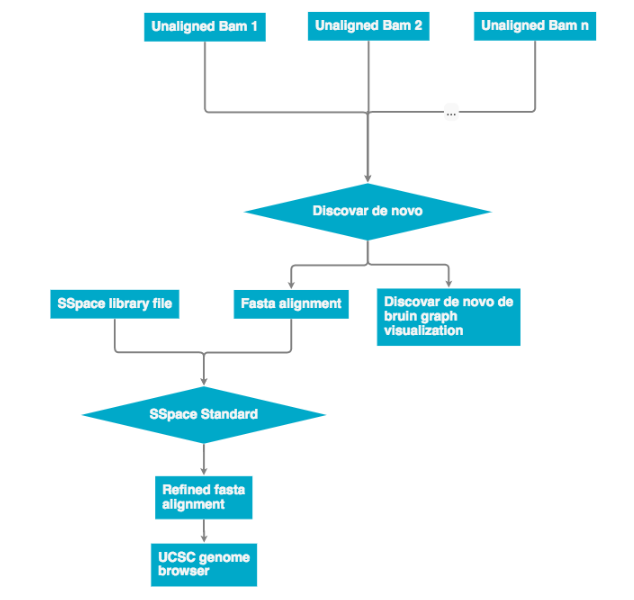
All unaligned BAM files are then passed into Discovar de novo. The output is an assembly in .fasta format and Discovar de novo visualization files. The .fasta file can then be re-scaffolded with a scaffolding program (see next workflow). The finished file can be represented on the UCSC genome browser.
Discovar de novo currently does not support mate pair libraries. To incorporate these data the program a program SSpace was used to re-scaffold the Discovar de novo fasta output. The workflow below describes how mate pair libraries are processed and factored into the assembly. The SSpace library is a small text file that points to the libraries and provided meta data statistics. Please see the 'Programs used' section for SSpace citation and further details.
FastQC of adapter-trimmed and PCR duplicate-removed data
After removing adapters and PCR duplicates, we run FastQC in two of the libraries. In general, the quality of the reads decrease in the last base-positions. Also, read 2 of the SW019 library shows problems in the per tile sequence quality. Bellow are the pdf files with the fastqc for the PCR and adapter removed libraries. The protocol we used to run fastqc is uploaded in this link: fastqc. The path to the files is: campusdata/gchaves/fastqc_trimmed_PCR_duplicates.
Fastq to bam
The fastq to bam conversion was performed using the picard toolset. Specifically the fastqToSam.jar file was used to prepare the bam files.
Discovar de novo run logs
Discovar de novo was designed for very specific data. To test the validity of our data we perform different test runs. The test runs used a percentage of data from the libraries available. All the tests were run on .bam files. The log files were run on edser2 or campusrocks nodes with more than 200 GB of RAM available. The run logs are stored as .txt files. The full logs can be seen on the wiki here,
| Run log | Data used |
| 1% data | (Pre Skewer and FastUniq) MiSeq data SW019_S1_L001, HiSeq data SW018_S1_L007, HiSeq data SW019_S2_L008 |
| 5% data | (Pre Skewer and FastUniq) MiSeq data SW019_S1_L001, HiSeq data SW018_S1_L007, HiSeq data SW019_S2_L008 |
| 10% data | (Pre Skewer and FastUniq) MiSeq data SW019_S1_L001, HiSeq data SW018_S1_L007, HiSeq data SW019_S2_L008 |
| 50% data | (Post Skewer and FastUniq) MiSeq data SW019_S1_L001, HiSeq data SW018_S1_L007, HiSeq data SW019_S2_L008 |
| 50% data UCSF | (Post Skewer and FastUniq) UCSF SW018 and SW019 data |
| Full data run 1 | (Post Skewer and FastUniq) 100% of the MiSeq SW019, UCSF SW019, and 50 % BS-tag datasets |
| Kolossus full run | (Post Skewer and FastUniq) 100% of the MiSeq SW019, UCSF SW019, UCSF SW018, BS-tag, BS-MK datasets |
The logs are very large, important statistics have been gathered and are compared below. Note that MPL1 is an acronym for mean length of first read in pair up to first error.
| 1% run | 5% run | 10 % run | 50 % run | 50% UCSF run | FullRun1 | Kollosus full run | |
| Total runtime | 1.75 hours | 1.53 hours | 2.4 hours | 8.53 hours | 14.9 hours | 24.2 hours | 103 hours |
| Peak memory use | 43.92 GB | 78.10 GB | 151.05 GB | 220.11 GB | 184.09 GB | 246.03 GB | 583.25 GB |
| Bases in 1kb+ scaffolds | 75,233 | 592,685 | 1,476,875 | 101,397,871 | 1,528,625,509 | 1,849,167,875 | 1,885,373,341 |
| Bases in 10kb+ scaffolds | 10,572 | 11,088 | 168,543 | 151,417 | 137,959,107 | 972,798,485 | 1,106,140,476 |
| MPL1 | 2 | 2 | 3 | 7 | 156 | 169 | 169 |
| Contig N50 | 2,622 | 2,067 | 2,563 | 1,489 | 3,979 | 9,513 | 10,427 |
| Scaffold N50 | 2,622 | 2,067 | 2,563 | 1,489 | 3,979 | 10,634 | 12,549 |
| Coverage | 16x | 47x | 80X |
Fasta assemblies
Our fasta assembly files are located at
/campusdata/BME235/S15_assemblies/DiscovarDeNovo
Each fasta assembly is in its own directory, the directory name is the assembly name. Currently there are five assemblies, The final fasta file is name a.lines.fasta. Note originally the authors used the a.fasta file (which contains the reverse complement of every contig) for statistics. Consequently statistics were falsely reported as twice the actual number until May 15th 2015 when the error was identified. The statistics have since been corrected.
Raw stats were mined from the fasta files using a python script fastaStats.py. The script is available online at (https://github.com/ChrisEisenhart/binfProgs/blob/master/basicProgs/fastaStats.py).
| Assembly name | Bytes | Total bases | # scafs | Av. scaf len | Longest scaf | Scaf N50 | # Scaf > 5Kb | Bases in 10kb+ scafs |
| 1%run | 463K | 448,486 | 2,558 | 350 | 5,385 | 2,622 | 10,572 | |
| 5%run | 3.4 M | 3,377,064 | 18,224 | 370 | 6,637 | 2,067 | 11,088 | |
| 10%run | 7.5 M | 7,382,612 | 38,195 | 386 | 11,911 | 2,563 | 168,543 | |
| 50%run | 137 M | 137,695,736 | 273,653 | 1,006 | 19,658 | 1,489 | 151,417 | |
| UCSF50%run | 1.9 G | 1,839,371,352 | 1,126,557 | 1,632 | 55,757 | 3,979 | 80,721 | 137,959,107 |
| firstFullRun | 2.2G | 2,245,788,654 | 1,450,447 | 1,548 | 153,999 | 10,634 | 118,545 | 972,798,485 |
| Kolossus full run | 2.4G | 2,395,797,282 | 1,843,153 | 1299 | 129,831 | 12,549 | 113,978 | 1,106,140,476 |
The absolute path to our latest assembly in .fasta format is;
/campusdata/BME235/S15_assemblies/DiscovarDeNovo/KolossusAssembly/discovarDeNovoKolossusAssembly.fasta
Looking at the 10% run, the majority of scaffolds generated are quite short (<1kb).
When a similar histogram is generated for the assembly made with 50% of the SW018 and SW019 reads from UCSF, you can see that the average scaffold length is higher and there are quite a few more scaffolds that are over 10kb.
Post assembly scaffolding
The program SSPace (documentation below) was used to scaffold the the assembly with mate pair data. The UCSF SW041 and SW042 mate pair libraries were used to generate the library.txt file.
BLAST results
The .fasta assemblies were run through BLAST. The results are below,
There seems to be a very high sequence identity with Notopygos (http://sv.wikipedia.org/wiki/Notopygos)
UCSC genome browser hub
See instructions for setting up the hub here, Banana slug browser
Programs used
The program, its location, and a brief, (brief!) explanation of what the program does
Picard
The Picard is a set of Java-based command-line utilities for SAM and BAM file manipulation (edser2:/soe/calef/picardtools and edser2:/soe/calef/picard_jars). Webpage: http://picard.sourceforge.net/.
Jemalloc
Is a general purpose malloc(3) implementation that emphasizes fragmentation avoidance and scalable concurrency support (edser2:/soe/calef/jemalloc). Webpage: http://www.canonware.com/jemalloc/.
Skewer
Skewer is an adapter trimmer for Illumina paired-end sequences (/campusdata/BME235/S15_assemblies/SOAPdenovo2/adapterRemovalTask/skewer_run). Webpage: http://sourceforge.net/projects/skewer/.
FastUniq
FastUniq is a fast de novo duplicate removal tool for paired short DNA sequences (/campusdata/BME235/bin). Webpage: http://sourceforge.net/projects/fastuniq/.
GCC
GCC is a compiler for the GNU operating system. Webpage: https://gcc.gnu.org/.
SSpace
A program for scaffolding fasta files, written by Marten Boetzer and Walter Pirovano. The authors are very protective over the use and citation of this program. The program can be obtained by at http://www.baseclear.com/genomics/bioinformatics/basetools/SSPACE.
Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W(2011), Scaffolding pre-assembled contigs using SSPACE, Bioinformatics 27(4):578-9
References
blog posts, related information etc.
http://blastedbio.blogspot.com/2011/10/fastq-must-die-long-live-sambam.html