Table of Contents
Banana slug genome browser
Currently the banana slug genome is supported as a private track hub. For those who know how to set up custom track hubs, the link is here. If it is your first time setting up a custom track hub, follow the instructions for 'Mounting the banana slug track hub' below!
http://hgwdev-ceisenhart.cse.ucsc.edu/~ceisenhart/hubs/bananaSlug/hub.txt
Currently the following assemblies are supported;
ABySS
SOAP
Discovar de novo
Mitochondrial
Mounting the banana slug track hub
To view the banana slug data please go to
https://genome.ucsc.edu/cgi-bin/hgGateway
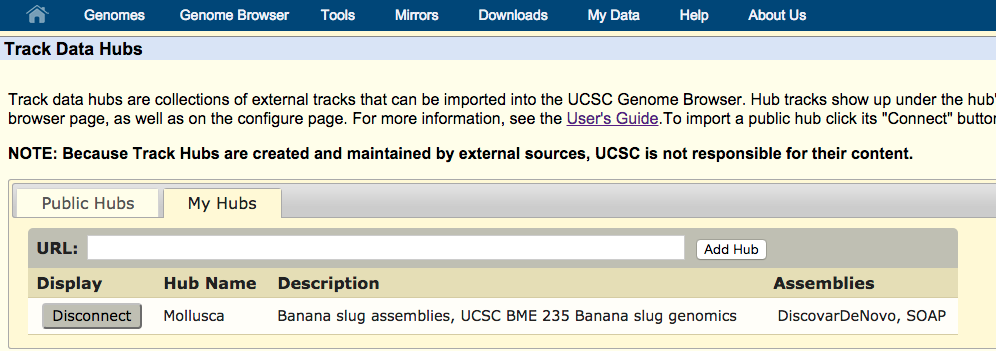
and click the 'track hubs' button
next navigate to the 'my hubs' tab
Paste the url provided,
http://hgwdev-ceisenhart.cse.ucsc.edu/~ceisenhart/hubs/bananaSlug/hub.txt
into the 'URL' text box and click the 'Add hub' button. You will be redirected to the banana slug hub which can be used as normal.
Creating a new track hub
To setup a custom track hub follow the instructions here.
Note the banana slug track hub has already been created. These are the instructions that were used to create it the first time, it can be accessed using the instructions above.