PreqC of adapter-trimmed and PCR duplicate-removed data
The initial datasets were ran through Skewer and FastUniq to create PCR duplicate free and adaptor free files. The libraries were condensed, so now the MiSeq and HiSeq 19 library are condensed. The information and location for these data are embedded in the library data pages. PreqC was ran on all these data combined.
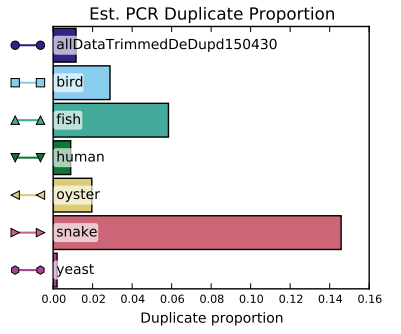 Note that the PCR duplication cannot be directly compared to the unprocessed data since the unprocessed data was ran for each library. By weighing the libraries based on file size a weighted PCR duplication percent for all the unprocessed files was calculated to be roughly 2.7%. This number can be directly compared to the percent duplication in these results above. It seems that the pre processing removed just under half of the total duplicates.
Note that the PCR duplication cannot be directly compared to the unprocessed data since the unprocessed data was ran for each library. By weighing the libraries based on file size a weighted PCR duplication percent for all the unprocessed files was calculated to be roughly 2.7%. This number can be directly compared to the percent duplication in these results above. It seems that the pre processing removed just under half of the total duplicates.
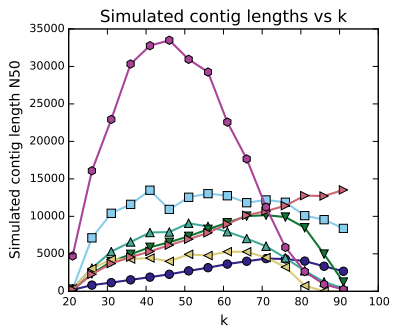 Additionally the ideal K-mer size for these data is longer than for the unprocessed data. This graph shows the ideal K-mer size to be around 75 bases. This is 15 bases longer than previous estimates.
Additionally the ideal K-mer size for these data is longer than for the unprocessed data. This graph shows the ideal K-mer size to be around 75 bases. This is 15 bases longer than previous estimates.

