Table of Contents
illumina Sequencing Run 1
Meta
Prepared by Akram Tariq Sial
Date:
Overview: This sample was prepared according to manufacturer's recommendations. [Add link to illumina protocol]
Starting DNA
Initial Size Selection
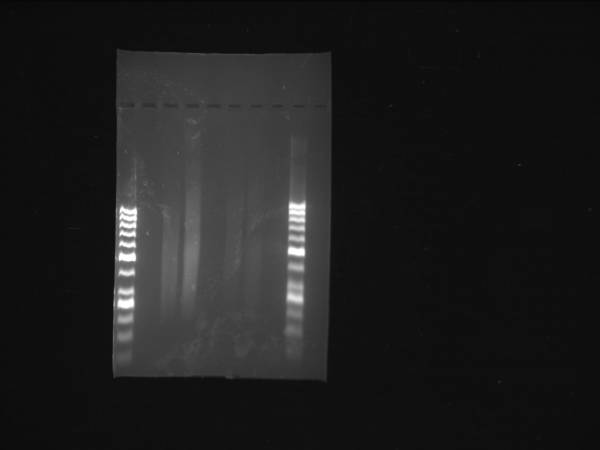
Fermentas O'generuler 50bp ladder used in these gels. Lanes 1 & 6
1µg of 98.0 ng/µl genomic DNA was sheared using either Covaris or a Nebulizer. Lanes 3&4: Banana Slug DNA sheared using Covaris. Lanes 5&6: Banana Slug DNA sheared using a Nebulizer. Both were cut between 250-375 basepairs.
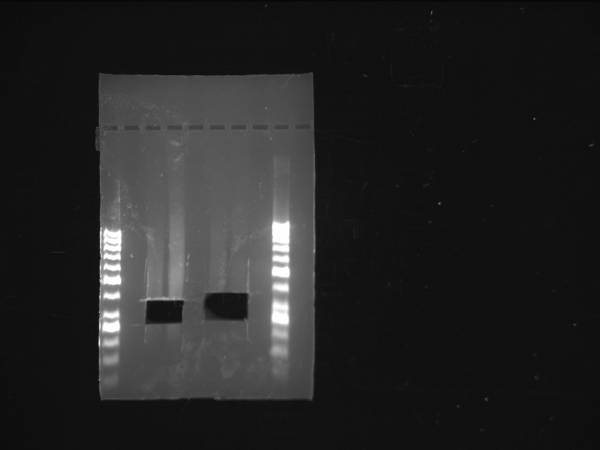
Bioanalyzer results of initial shearing and final amplified library size selection
Bioanalyzer results of shearing distributions and final amplified libraries. Covaris and Nebulizer shearing are in lanes 1 and 2 respectively. Amplified and size selected libraries for Nebulizer and Covaris in lanes 3 and 4 respectively.
The results indicate the final nebulizer library ranges from 202-320bp in distribution. The final covaris library ranges from 225-263bp in length.
Final distribution of paired end reads
Each stage indicates a different distribution in the length of the paired-end reads. The original size selection cut indicates a fragment size of 250-375. However, the bioanalyzer results indicate the final size selected fragment is much shorter at 200-320 bp for a final distribution between 100-230 bp.
Differences between lanes in illumina run
Each lane contained a different concentration of starting material used in bridge amplification.
| Lane | Starting amount of library and library type |
|---|---|
| Lane 1 | 8 picoMoles Nebulized Library |
| Lane 2 | 10 picoMoles Nebulized Library |
| Lane 3 | 12 picoMoles Nebulized Library |
| Lane 4 | 6 picoMoles phix control* |
| Lane 5 | 8 picoMoles Covaris Library |
| Lane 6 | 10 picoMoles Covaris Library |
| Lane 7 | 12 picoMoles Covaris Library |
| Lane 8 | 14 picoMoles Covaris Library |
These different starting quantities of DNA are reflected in the number of clusters generated and the total reads outputted for each lane. Additionally, a different distribution of distance for the paired-end reads are different for lanes 1-3 and 5-8
*Data from lane 4 of the illumina run SHOULD NOT be used for assembly