Table of Contents
Banana Slug RNA-Seq
To see how we are using this data, please view this page.
Raw data
The raw data can be accessed online here.
Total = 56G Raw unprocessed data
FastQC
The fastQC reports for the raw files can be found online here raw fastqc reports.
Processed data
fastUniq fastUniq removes PCR duplicates.
fastUniq + fastqQuality fastqQuality removes low quality reads.
FastQC reports for the processed files can be found in the 'fastQC' subdirectories.
RNA Extraction
RNA was extracted from three tissues, the albumen, proximal albumen and penis. The protocol used for RNA extraction is as follows:
Sample Crushing
- Place sample tubes on dry ice.
- Place receiving tubes on dry ice.
- Fill cleaned mortar + pestle with liquid nitrogen.
- Wait until liquid calms down and then add more liquid nitrogen as well as one of your samples.
- Crush the sample into a fine powder.
- Once liquid nitrogen has reduced to small puddle, tilt the mortar onto its side.
- Once all liquid is gone, immediately place the powder into a receiving tube and place it back on dry ice.
- Clean scoop, mortar and pestle and repeat for all samples.
- Place samples in -80 Celsius.
Cleaning Procedure for Mortar and Pestle
- Rinse them with tap water.
- Add bleach to the bottom of the mortar then fill with water and let the mortar and pestle sit for 2 to 3 minutes.
- Rinse with water until bubbles do not appear.
- Dry with towels.
- Spritz mortar and pestle with RNAseZap and DNAzap.
- Dry with wipes.
- Spritz with 70% ethanol.
- Dry with wipes.
- Rinse out with RNA-free water.
- Dry with wipes.
Cleaning Procedure for Scoop
- Spritz with RNAseZap + DNAzap.
- Dry with Wipes.
- Spritz with 70% ethanol.
- Dry with Wipes.
RNA Isolation
This is basically a phenol-chloroform extraction followed by a Qiagen RNAeasy Mini Kit extraction.
- Add 1mL of TRIzol per 100mg of tissue
- Using a syringe,homogenize the sample by forcing the tissue pellets through the needle and back into the solution.
- Add 0.2ml of chloroform per 1ml of TRIzol and shake tube for 20 seconds
- Incubate for 3 minutes.
- Centrifuge the sample at 12,000xg for 15 minutes at 4 degrees Celsius.
- Remove the aqueous phase of the sample and place into a new tube.
- Adjust the sample volume to 100 microlitres with RNA-free water.
- Add 350 microlitres of RLT Buffer and mix well.
- Add 250 microlitres of pure ethanol and mix well.
- Transfer solution to mini spin column and collection tube and centrifuge for 15 seconds and 9,000xg and discard flow-through.
- Add 500 microlitres Buffer RPE to the spin column and centrifuge at 9,000xg for 15 seconds and discard flow-through.
- Add 500 microlitres Buffer RPE to the spin column and centrifuge at 9,000xg for 2 minutes and discard flow-through.
- Place the spin column in a new collection tube and centrifuge it for 2 minutes at 15,000xg.
- Place the spin column in a new collection tube. Add 50 microlitres of RNA-free water and centrifuge for 1 minute and 9,000xg to elute the RNA.
- Measure the concentration and run a portion of the RNA on a gel to check quality.
- Immediately place in -80 degrees Celsius.
Results of RNA Extraction
- The albumen produced 2055 ng/ul of RNA in 50ul of elution.
- The proximal albumen produced 493.7 ng/ul of RNA in 50ul of elution.
- The penis produced 78.28 ng/ul of RNA in 50ul of elution.
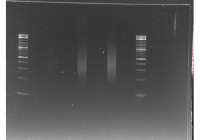
 The contents of the gel image are as follows:
The contents of the gel image are as follows:
- The first column is the ladder.
- The second column is the albumen.
- The third column is the proximal albumen.
- The fourth column is the penis.
- The rest of the columns are unrelated B cell RNA extractions.
The differences in the intensity of the bands are probably due to the wildly different concentrations of RNA in the elution. The pattern in the B cell columns is what is more typical for RNA extractions. However, if the RNA was heavily degraded you would expect to see a very strong smear of short RNAs near the bottom. Thus, I would concluded that this RNA is probably good enough to be used for RNA-Seq.
cDNA Synthesis
Reaction Mixture 1
- 2 ul RNA at ~ 20ng/ul.
- 1 ul oligo-dT (10um).
- 1 ul dntp (10um).
Reaction Protocol 1
- 72 degrees Celsius for 3 minutes.
- Immediately place on ice for 2 minutes.
Reaction Mixture 2
- 4 ul Reaction Mixture 1.
- 0.5 ul superscript 2.
- 0.25ul RNAse inhibitor.
- 2 ul 5xFS buffer.
- 0.5 ul DTT.
- 2 ul betaine.
- 0.06 ul mgcl2.
- 0.1 ul 18N_Smartseq TSO.
- 0.59 ul H2O.
Reaction Protocol 2
- 42 degrees Celsius for 90 minutes.
- 70 degrees Celsius for 5 minutes.
- 4 degrees Celsius hold.
Reaction Mixture 3
- 10 ul Reaction Mixture 2.
- 12.5 ul Kappa Hifi Hotstart mix.
- 0.125 ul FS PCR primers(10um).
- 1.125 ul 1:10 RNAse A.
- 1.125 ul 1:10 Lambda Exonuclease.
Reaction Protocol 3
- 37 degrees Celsius for 30 minutes.
- 95 degrees Celsius for 3 minutes.
- 27 cycles of
- 98 degrees Celsius for 20 seconds.
- 67 degrees Celsius for 15 seconds.
- 72 degrees Celsius for 4 minutes.
- 72 degrees Celsius for 5 minutes.
- 4 degrees Celsius hold.
Purification Protocol
- Mix equal volumes SPRI bead mix and reaction mixture in a well plate and let sit for 2 minutes.
- Place the plate on the magnet aperture and wait for the mixture to completely clear.
- Remove the clear liquid.
- Wash twice with 120ul of 70% ethanol
- Let the beads dry for two minutes.
- Remove the plate from magnets and mix 30 ul TE buffer with beads and let sit for 2 minutes.
- place the plate back on the magnets and wait until the mixture clears.
- Elute the clear liquid and place in a tube. This elution contains the DNA.
cDNA Synthesis Results
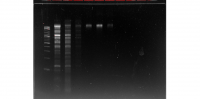
 The contents of the gel image are as follows:
The contents of the gel image are as follows:
- The first column is the ladder.
- The second column is the albumen cDNA.
- The third column is the proximal albumen cDNA.
- The fourth column is the penis cDNA.
- The fifth column is the proximal albumen cDNA done in parallel.
- The sixth column is the penis cDNA done in parallel.
- The seventh column is the albumen cDNA done in parallel.
The cDNA libraries done in parallel were created using a slightly different protocol which I don't have at the moment. This image indicates that the cDNA synthesis reactions and purifications were successful. There is an abundance of large cDNAs in the proximal albumen and penis libraries. In addition, there appear to be very few primer dimers.
Tagmentase Protocol
Tagmentase Prep
- Combine equal volumes of Tn5 forward and reverse oligos
- Heat oligo mixture to 95 degrees Celsius.
- Cool to the mixture to room temperature at 1 degree Celsius per second
- Add 1.5:10 volumes of a 100 um eqiumolar mixture of pre-annealed oligos to the Enzyme
- Incubate at room temperature for 60 minutes.
Transposase Mixture
- 11 ul H2O
- 5 ul DNA at ~ 50ng/ul
- 1 ul incubated TN5
- 4 ul TAPS PEG buffer
Transposase Reaction
- Incubate at 55 degrees Celsius for 7 minutes
- Add 5 ul of 0.2% SDS
- Incubate at room temperature for 7 minutes
Amplification Mixture
- 12.5 ul 2x Kappa hifi hotstart mix
- 1 ul TN5ME-A primer
- 1 ul TN5ME-B primer
- 5.5 ul H2O
Amplification Protocol
- 98 degrees Celsius for 3 minutes
- Add 5 ul of the transposase reaction mixture
- 72 degrees Celsius for 6 minutes
- 98 degrees Celsius for 30 seconds
- 8 cycles of:
- 98 degrees Celsius for 10 seconds
- 63 degrees Celsius for 30 seconds
- 72 degrees Celsius for 2 minutes
Library prep results
After purifying the mixtures as described previously, they were run on a gel and size selected. The DNA was then purified using a standard gel extraction protocol. The results are as follows:
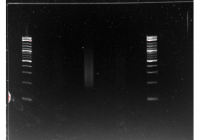
This gel image shows the size range of the tagmentase libraries for the Albumen, Proximal Albumen and Penis in that order. The Albumen was too faint to see so it was redone. The other two tissues were size selected for everything over 300bp in size. The gel extractions for each tissue was eluted in 40ul of TE buffer.
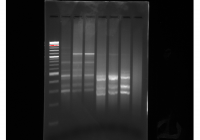
This gel image shows redone triplicates of the Albumen tissue libraries. The central library appeared to have the greatest concentration of DNA so it was size selected for everything over 300bp. The gel was eluted in 40ul of TE buffer.
RNA-Seq library testing
All six libraries were sequenced at low concentrations on a MiSeq platform. Preliminary data can be found on campusrocks at /campusdata/BME235/Spring2015Data/RNA_Seq/Preliminary_data
- The folder BS_P-25598754 contain the Penis tissue data.
- The folder BS_PA-25598752 contains the Proximal Albumen tissue data.
- the folder BS_AL-25598753 contain the Albumen tissue data.
Adapters for the libraries can be found at /campusdata/BME235/Spring2015Data/RNA_Seq/Preliminary_data/Banana_Slug_Pool_6-4-2015.tsv
Also, Ashley's reads will have a TSO at the beginning of the forward read. This needs to be removed before analysis can continue. The TSO sequence is “GCCTAAGCCATCGGG”